Definition of recombinant DNA:
Tools and steps that are used in recombinant DNA technique:
- first of all enzymes are required for the manipulation of DNA molecule
- vector which will be required for gene cloning.
- than the process of cloning of rDNA
- next step is the characterization of the gene which was cloned.
- than is the construction of genomic library.
- than is the application of rDNA technology.
History of recombinant RNA technology
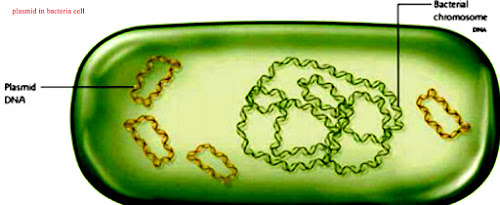
- in different plasmid the traits that are encoded the replication regions are mostly not essential for bacterial cell.
- mostly antibiotic resistant traits are present on these plasmids.
- in 1971, Cohen, a scientist, exploited the plasmid’s antibiotic resistance nature to the selectively enrich off spring that have cell propagating plasmid.
- than in the late 60s, it was observed that CaCl2 makes the E. Coli permeable for the DNA molecule but the E. Coli cell was not able to grow with this changed genetic makeup.
- than in 1972, Berg came up with the technique of joining fragments of DNA in outside of the cell using enzyme ligase for this process.
- for the purpose of cutting DNA endonuclease and restriction enzymes are used.
- than in 1972, Berg, Cohen and Boyer met up for the discussion of a collaborative project so that after that they would be able to start up the process of recombinant DNA technology.
- than in the month of March in 1973, they produce DNA fragments using the technique of Boyer and than join them with the plasmid using Berg’s method and than inserted it into the host which is the bacteria cell using Cohen’s method.
- in this way the first demonstration of the technique how DNA cloning take place was achieved.
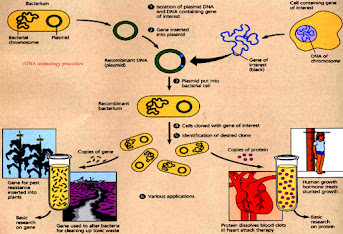
Recombinant DNA technology procedure:
- first of all plasmid DNA and the DNA containing gene of interest is isolated.
- than gene is to be inserted into the plasmid.
- than the genetically altered plasmid is inserted into the host which is bacterial cell.
- than cloning of the cell containing gene of interest take place
- than the desired gene is identified.
Essential elements of rDNA technology:
- cutting enzymes i.e. that are restriction endonucleases
- joining enzymes i.e. that are ligases
- and the vectors that are used for gene cloning i.e. plasmid
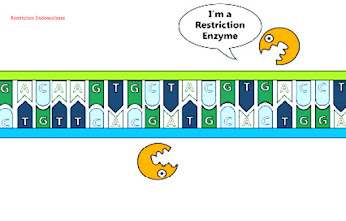
Restriction endonucleases:
- these enzymes are also called endonucleases.
- these restriction enzymes are present in bacteria for cutting DNA molecule ay the specific sequence
- its main biological function is to destroy fully the foreign invading DNA molecule in bacterial cell
- these restriction enzymes are named after the organism from which these enzymes obtained.
- for example : like EcoRI is the restriction enzyme obtained from E. Coli strain RY13.
- BamHI is another example which is derived from Bacillus amyloliquefaciens.
- bacteria protect its own DNA from the action of these cutting enzymes by methylation on specific sequence of DNA.
Types of restriction enzyme:
- these can of any type from; type-1,type-II,type-III, and type IV.
- most commonly available and commercially used restriction enzymes are of type II i.e. more than 400 enzymes available.
- Palindromic sequence which is of 4-8 nucleotide is their recognition site.
- restriction enzyme may produce blunt or sticky ends.